Overview
The overarching goal of the laboratory is to unravel the role of telomere dysfunction in gliomagenesis and evolution. While the biochemistry of telomere dysfunction has been defined at a high resolution, little is known about its broader impact on tumor genomes and role in tumorigenesis and evolution. The laboratory will map the full ramifications of telomere dysfunction on glioma genomes, both as a tumor suppressor by promoting genomic instability, and as an oncogene by facilitating tumorigenic mutations. To achieve these goals, we employ custom next-generation genomics technologies (whole genome, long read, chromatin capture and single-cell sequencing) on rigorously engineered model systems and patient samples.
Figure 1. Tumor phylogenetic trees (ancestry trees) linking multiple samples from three distinct gliomas projected into its original spatial context and onto a reference brain space. Samples from each tumor are shown by labeled points and the phylogeny (ancestry tree) was transformed in phylomorphospace using the sample coordinates as parameters. Lines represent phylogenetic (ancestral) relationships between samples. Tumor volumes are indicated by the gold (FLAIR) and orange-red (T1+Gado) overlays. Subcortical structures are shown as colored volumes. A reference brain volume (Montreal Neurological Institute) is shown in gray.
Research Projects
Genomic dissection of structural mutations following telomere dysfunction in glioma
Glioma genomes are littered with somatic structural variants, but the mutational processes that give rise to these events are poorly understood. Repetitive DNA elements called telomeres make up chromosome termini. Their function, mediated by the protein complex shelterin, is to prevent endogenous DNA repair processes from engaging in the same manner that they would engage double strand breaks. Dysfunctional telomeres lack this end-protection and become hotspots for structural variation. We hypothesize that telomere dysfunction gives rise to some of the characteristic variation observed in glioma. Our lab is using long and short-read sequencing to dissect the structural genomic signatures of artificially induced and spontaneous telomere dysfunction in isogenic cell line models. We are extrapolating these patterns to publicly available sequencing datasets. Disentangling the vast landscape of structural mutations in glioma and reconstructing their origin will revise our conceptual understanding on the origin of disease.
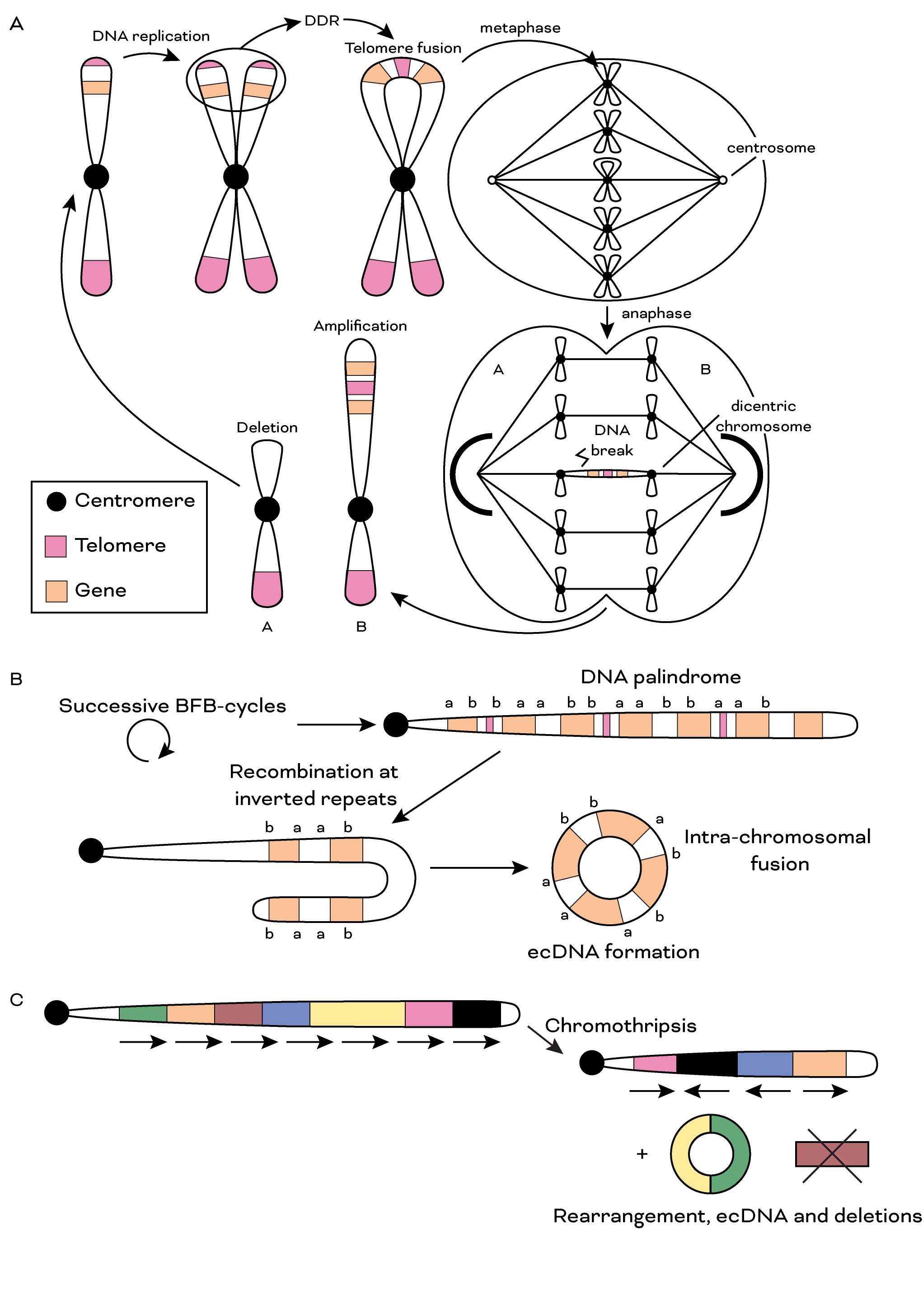
Figure 2. Genomic instability related to telomere dysfunction. a. Schematic illustrating BFB-cycles. Following a single BFB-cycle, daughter cells are left with unequal DNA content, leading to unequal seggregation of genetic material. BFB-cycles may also involve fusion of non-sister telomeres (not shown). b. Successive BFB-cycles may form DNA palindromes demonstrating high intra-segmental homology. This can lead to intra-chromosomal fusions and formation of double minutes. c. Telomere dysfunction has also been linked to chromotripsis and following this phenomenon DNA segments can be rearranged, lost or circularized. DDR = DNA damage response; BFB = breakage-fusion-bridge
Reconstructing evolutionary trajectories of glioma
Sustained telomere dysfunction drives tumor genomes to the brink of destruction. Yet somehow cancer genomes seem to thrive under these catastrophic conditions. Tumor development can be thought of as an evolutionary process. Advantagous cellular changes promoting fitness are under selection to survive, whereas cellular lineages acquiring disadvantageous change rapidly become extinct. Using deep DNA and single-cell sequencing we study telomere dysfunction through the lens of evolution, piecing together distinct cellular lineages and temporally arranging genomic variation that arises from telomere dysfunction and other mutational processes. In reconstructing the evolutionary trajectories of tumorigenesis and progression we aim to identify targetable evolutionary bottlenecks and therapeutic vulnerabilties.
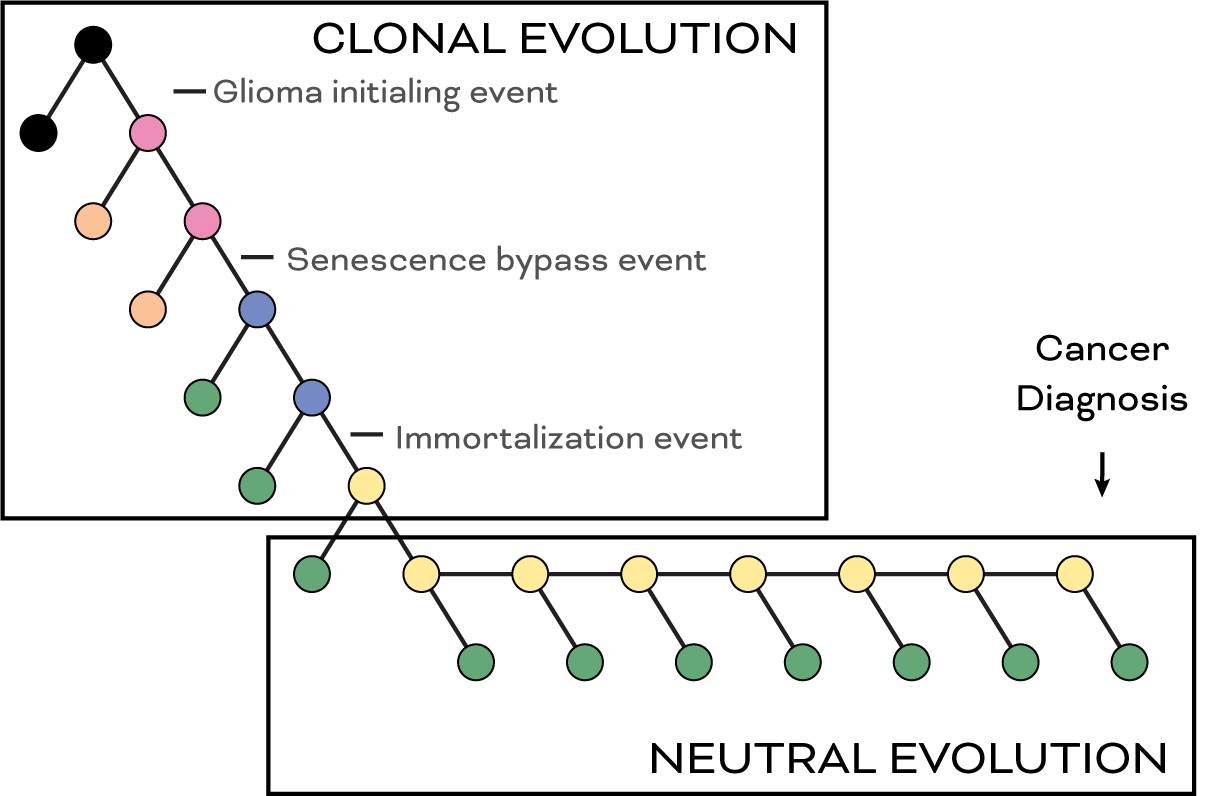
Figure 3. Clonal evolution (selection) and neutral evolution (drift) both contribute to glioma development. Variation due to telomere dysfunction may fit either category.
Defining the regulatory landscape of functional and dysfunctional telomeric chromatin interactions
Interphase telomeres form long-range chromatin interactions with other elements of the genome, but it is unclear what purpose these interactions serve. It was shown that telomeric sequences repress the transcription of nearby genes in a phenomenon known as the “telomere position effect”. Moreover, distal elements called “enhancers” can interact with a gene’s proximal promoter to regulate its transcription. Our laboratory is investigating whether telomeric chromatin interactions can interfere with enhancer-promoter interactions to play a hand in transcriptional regulation. We have developed a telomere interaction assay called “Telomere-C” to identify interactions between telomeres and the rest of the genome. Through employing Telomere-C in tightly controlled cellular systems, we will resolve whether telomeric chromatin interactions can control transcriptional permissiveness.