The Barthel laboratory is focused on unraveling the role of telomere dysfunction in glioma development and evolution. We employ computational biology, evolutionary genetics, functional genomics and molecular biology to map the ramifications of dysfunctional telomeres on glioma genomes. We are looking for a talented, passionate and highly motivated individual to enrich our team and study telomere dysfunction.
Position Summary
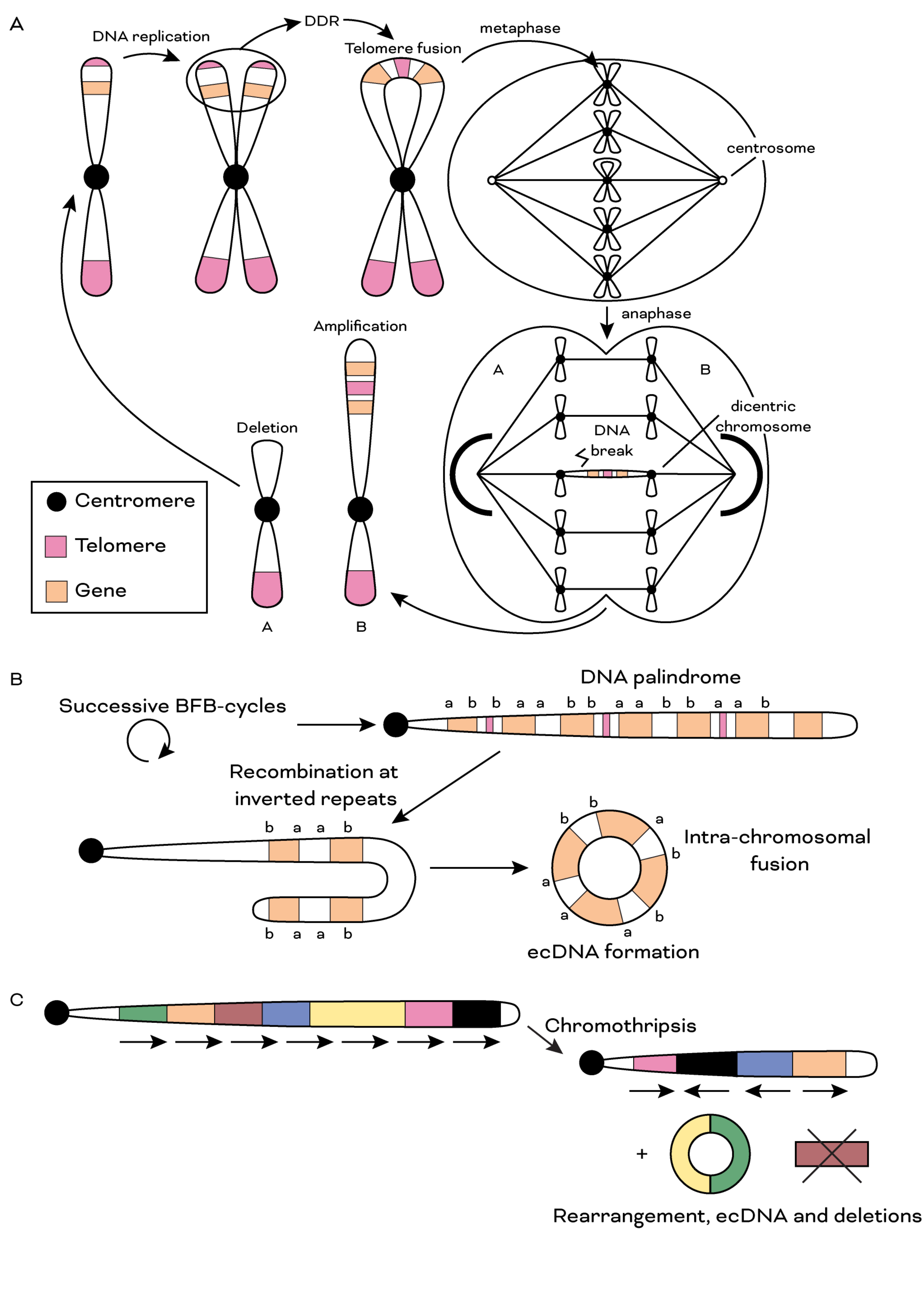
This project concentrates on detecting telomere dysfunction from DNA sequencing. While the biochemistry of telomere fusions has been described in detail, the genomics of this phenomenon remain largely a mystery, predominantly due to its repetitive sequence and origin in uncharted regions of the genome. The successful candidate will develop algorithmic solutions to dissect signatures of telomere dysfunction from long and short read sequencing. They will be involved in data analysis, interpretation and write manuscripts to describe their findings. They will collaborate with bench scientists to validate their findings using traditional approaches and develop in vitro models to test new hypotheses. They will lead their own project but will be a part of a team and help on other projects as well.
Career Development
We value our trainee’s personal development and strive to unlock their full potential. Trainees are provided with ample opportunities for additional training in computational and cancer biology. Successful applicants are encouraged to attend scientific meetings and the lab will provide support for at least one scientific conference per year. While this position is not contingent upon securing additional funding, successful candidates will be supported and encouraged to apply for outside sources of funding. Candidates are welcomed to explore their own scientific interests into areas of research that fall within the general scope of the lab.
Education and Preferred Qualifications
The candidate must have experience in Unix-based computing environments with a demonstrated knowledge of dynamic scripting languages such as Python or Perl. A fundamental understanding of low-level programming languages such as C/C++/Java/Rust/Go is preferred. A basic understanding of statistics is a plus. The ideal candidate will have peer reviewed publications utilizing DNA sequencing. An emphasis will be placed on candidates with a track record of method development. An understanding of cancer biology and genomics is a plus. Candidates of all backgrounds will be considered.
Location
We are located at the Translational Genomics Institute (TGen) set to the backdrop of the stunning Sonoran Desert. The successful candidate will be a part of the Cancer and Cell Biology Division at TGen and will have numerous opportunities to work with our partners around the globe.
Application
Please direct all questions to barthel@barthel-lab.com. Application via the “Apply” button on the left menu. Attach a letter describing your research interests and motivation to join our group. Include an academic CV detailing your past research experience, publication track record and (conference) presentations. We will start reviewing applications immediately and until the position is filled.